|
Introduction
Two
million people in India die each year due to
infectious diseases [1]. Septicaemia or sepsis
results when circulating bacteria in blood
multiply at a rate that surpasses their
elimination by phagocytes [2]. Blood infections
are a substantial reason for morbidity and
mortality of patients, particularly in developing
countries [3]. If left untreated, bloodstream
infections may lead to more dangerous infections,
involving all organs and ultimately death [4].
Bloodstream infection (BSI) is a leading cause of
mortality in critically ill patients. Clinical
microbiology laboratory performs identification
(ID) and antimicrobial susceptibility testing
(AST) to guide antibiotic therapy and possible
drug resistance. Rapid bacterial identification
and susceptibility testing improve patient therapy
and outcome, decreases emergence of resistance
[5]. There is a need to provide rapid, efficient
and accurate system for identification and
antimicrobial susceptibility testing of pathogens.
In this regard the automated identification/AST
systems aid in rapid diagnosis/treatment of
bacterial pathogens [6]. Automated blood culture
systems and automated identification and
susceptibility testing of bacteria have been in
the market for a number of years however
application of automated systems in Microbiology
is different than other clinical laboratories [7].
Although classical
identification methods are still considered the
gold standard, these methods are slow, time
consuming and prone to subjective interpretations.
On the other hand, the Vitek-2 compact system
reduces the time necessary for identification and
permits the standardization of inter- and
intra-laboratory results, the storage of results,
issuing rapid epidemiological reports, and
simultaneous identification and antimicrobial
susceptibility testing; however, the system is
poorly efficient in identifying certain species of
Gram-positive cocci [8].
Automated systems
use sophisticated software to analyse the growth
rates and determine the antibiotic minimum
inhibitory concentration (MIC) for the organism by
using specialized decision technology. Although
there are differences among each system the
general process of identification is almost same.
The Vitek-2 compact system is the second
generation of Vitek-2 compact system and offers a
more sophisticated model of data analysis as well
as a fully automated process for card
identification, organism suspension dilution, and
card filling [9]. Nonetheless a reduction in
labour, faster reporting of results especially in
blood stream infections and identification of
uncommon or rare organisms are some of the reasons
as to why automation in microbiology has gained
popularity in recent years [10]. The current study
was thus designed to evaluate the Vitek-2 compact
system with conventional blood culture method from
flagged BacT/ALERT blood culture bottles causing
neonatal septicaemia in a tertiary care hospital.
Materials and Methods
This retrospective study was conducted in the
Department of Microbiology at University College
of Medical Sciences and GTBH, New Delhi over a
period of one year from January 2023 to January
2024. A total of 170 non duplicate isolates of
both Gram positive and Gram-negative organisms
recovered from blood samples were included.
Sample Processing: After
receiving the blood sample of the neonate of
suspected septicaemia in the laboratory, the
bottles were checked for adequacy of volume and
labelling errors. The conventional bottles were
processed accordingly as described below.
(a)
Identification by conventional method: Phenotypic
identification consisted of Gram staining for the
observation of morphology and specific staining,
followed by a series of biochemical tests specific
for each group of microorganisms. Gram-positive
cocci were submitted to the catalase test for
differentiation between Staphylococcus
and Enterococcus. The following
biochemical test battery was used for the
identification of species of the genus Staphylococcus:
coagulase, sugar fermentation (sucrose, maltose,
trehalose, xylose, and mannitol), anaerobic growth
on semi-solid sodium thioglycolate medium and, if
necessary, ornithine and urease production and
novobiocin susceptibility [11]. Isolates
previously identified as Gram-positive,
catalase-negative, bile esculin-positive,
NaCl-positive (growth in brain heart infusion
broth with 6.5 % NaCl) and pyrrolidonyl
aminopeptidase test-positive cocci were submitted
to biochemical tests of fermentation of mannitol,
arabinose, arginine and sorbitol, motility, and
presence or absence of a pigment on sheep blood
agar. Gram-negative bacilli were first tested for
glucose fermentation. Glucose-fermenting bacilli
were submitted to manual biochemical tests known
as Citrate, an identification system based on the
following tests: production of H2S, urease and
l-tryptophan desaminase; motility; Indol
production; lysine decarboxylase production, and
the ability to use citrate as a single carbon
source. Non-glucose-fermenting Gram-negative
bacilli were identified based on motility, growth
at a temperature of 42 °C, and production of
DNAase.
(b)
Automated identification: Samples
exhibiting microbial growth were submitted to Gram
staining and cultured on solid media directly from
the blood culture bottles. After initial growth on
blood agar (for Gram-positive cocci, Gram-positive
bacilli, and yeast) and MacConkey agar (for
Gram-negative bacilli), colonies were sub-cultured
to ensure purity and then inoculated into specific
identification cards for the VITEK-2 system.
Gram-positive cocci, Gram-positive bacilli and
yeast were inoculated into the cards from colonies
grown on blood agar and Gram-negative bacilli from
colonies grown on MacConkey agar, all diluted in
saline (0.9 % NaCl) to a 0.5 McFarland standard.
The VITEK-2 compact system automatically processes
the inoculated cards. Each card contains a variety
of biochemical tests that help identify the
species. The system interprets the results based
on the growth and biochemical reactions observed,
providing a detailed report that includes the
identification of the microorganism.
Quality
Control: Staphylococcus aureus
ATCC 25923, Enterococcus faecalis ATCC
29212, Escherichia coli ATCC 25922, Pseudomonas
aeruginosa ATCC 27853, and Klebsiella
pneumoniae ATCC 700603 were used as quality
control (QC) standard strains for both methods.
Statistical
Analysis: The results were compared by
entering the data on excel sheets and simple
statistical calculations were made and recorded.
Results
A total of 170 non
duplicate isolates of Gram positive and
Gram-negative bacteria recovered from the blood
samples of the patients admitted at GTBH were
included in the study. Gram negative organisms
were recovered from 97 isolates (57.1%) and 73
isolates (42.9%) were gram positive organisms
which included all the samples received from the
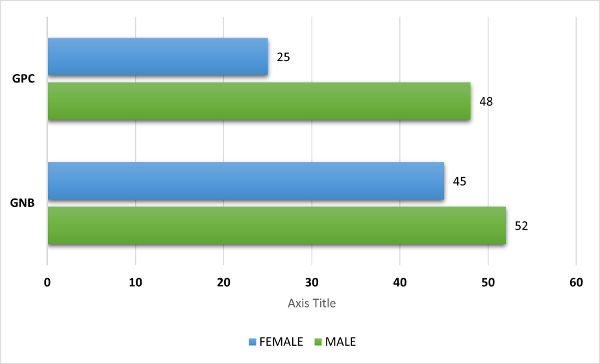
neonatal age group [Figure 1]. Patients from whom
Gram negative isolates (97) were recovered
included 52 (53.6%) males and 45 (46.3%) females.
Most of the Gram-Negative bacteria identified
included Klebsiella pneumoniae (n=47;
48.4%) followed by Acinetobacter baumannii
(n=15; 15.4%), Enterobacter cloacae (n=14;
14.4%), Citrobacter species (n=10;
10.3%), Pseudomonas aeruginosa (n=05;
5.1%), Escherichia coli (n=04; 4.4%) and
Burkholderia cepacia (n=02; 2.0%) [Table
1].
|
| Figure
1: Distribution of isolates of GPC and GNB
|
Concordant
identification (ID) results of Vitek-2 when
compared to the manual methods were seen with all
the isolates of Enterobacter cloacae,
Pseudomonas aeruginosa and
Burkholderia cepacia. However, discrepancy
in results of Vitek-2 compact system versus
conventional methods was seen for 2 isolates of E.
coli which were Acinetobacter
baumannii complex by Vitek-2. Likewise, 3
isolates of Klebsiella pneumoniae were
identified by Vitek-2 as Klebsiella oxytoca.
Three (3) isolates of Citrobacter species
were identified as E. coli by Vitek-2
compact system. In addition, 2 isolates of Acinetobacter
baumannii were identified as Enterobacter
cloacae in the Vitek-2 compact system. In
case of gram-positive organisms (73), mostly were
male patients. They are identified as Coagulase
negative staphylococcus (n=38; 52.05%)
followed by Staphylococcus aureus (n=27;
36.9%), Enterococcus faecalis (n=05;
6.8%) and Enterococcus faecium (n=03;
4.1%). Concordance between the isolates
obtained from Vitek-2 compact system and manual ID
was seen with all the isolates of Enterococcus
faecium and Enterococcus faecalis. However
discordant results were obtained for CONS and Staphylococcus
aureus, as 3 isolates of CONS and 6
isolates of Staphylococcus aureus were
identified as Enterococcus faecalis [Table
2].
|
Table 1: Gram negative organisms
identified by Vitek-2 compact system in
the study population (n=97)
|
|
Organism
|
Number of isolates N=97
|
Percentage (%)
|
Concordant
|
Discordant
|
|
Klebsiella species
|
47
|
48.4%
|
44
|
03
|
|
Acinetobacter baumannii
|
15
|
15.4%
|
13
|
02
|
|
Enterobacter cloacae
|
14
|
14.4%
|
14
|
0
|
|
Citrobacter species
|
10
|
10.3%
|
07
|
03
|
|
Pseudomonas aeruginosa
|
05
|
5.1%
|
05
|
0
|
|
Escherichia coli
|
04
|
4.4%
|
02
|
02
|
|
Burkholderia cepacia
|
02
|
2.0%
|
02
|
0
|
|
Totals
|
97
|
100%
|
87
|
10
|
|
Table 2: Gram positive organisms
identified by Vitek-2 compact system in
the study population (n=73)
|
|
Organism
|
Number of isolates N=63
|
Percentage (%)
|
Concordant
|
Discordant
|
|
CONS
|
38
|
52%
|
35
|
03
|
|
Staphylococcus aureus
|
27
|
36.9%
|
21
|
06
|
|
Enterococcus faecalis
|
05
|
6.8%
|
5
|
0
|
|
Enterococcus faecium
|
03
|
4.1%
|
3
|
0
|
|
Total
|
73
|
100%
|
64
|
09
|
Discussion
The need for the
rapid and efficient identification of
microorganisms isolated from blood cultures has
encouraged studies that investigated automated
identification systems to reduce the time of
identification. The early identification of
bloodstream infections allows for the early
modification of antimicrobial treatment and a
diminished need for other diagnostic tests. Thus,
patients’ hospital stay can be shortened, and
expenditures on patients can be reduced. Several
of these studies have used direct inoculation from
blood culture bottles, but the results were not as
efficient as those obtained in studies using
standard inocula from subcultures of
microorganisms grown for 24 hours on solid media.
Rapid bacterial identification and susceptibility
testing improve patient therapy and outcome,
decreases emergence of resistance and also reduces
costs [10].
The present study
included a total of 170 isolates from blood
samples, out of which 97 isolates were gram
negative and 73 were gram positive. In another
comparative study done by Donay JL et al [12],
evaluating the identification and antimicrobial
susceptibility testing performances of the BD
Phoenix Automated Microbiology System, a total of
305 clinical isolates were included, out of these
187 were Gram negative and 118 were Gram positive.
Samples were received more from male patients in
our study, similar to the previous study done by
Nadheema et al [13].
Among the 73
isolates of gram-positive cocci obtained, Staphylococcus
epidermidis (Coagulase negative
staphylococcus) followed by Staphylococcus
aureus were the most common Gram-positive
organism in our study. Similar results were seen
in previous studies done by Akgun et al [14] that
reported 71 (61.2%) as coagulase-negative Staphylococcus
(CoNS), 7 (6.0%) as Staphylococcus aureus
while 20 (17.2%) were Enterococcus
species. Also, another study done by Lupetti et al
[15]. reported the CoNS to be slightly lower than
S. aureus (62%). Another study by Chen et
al [16] reported that out of 197 (79.4%) isolates
of 248 Gram-positive organisms, 95 isolates were
CoNS and 58 isolates were S. aureus.
Out of the 97
gram-negative rods studied, 75 (77.5%)
corresponded to the family Enterobacteriaceae
(47 Klebsiella species, 14 Enterobacter
cloacae, 10 Citrobacter species
and 04 Escherichia coli) and 20 (20.4%)
were identified as non-fermentative gram-negative
rods (15 Acinetobacter baumannii and 5
Pseudomonas aeruginosa). In our study Klebsiella
species followed by Acinetobacter
baumannii were the most common
Gram-negative pathogen. Similar results were seen
in a study done by Jagadish et al. [17]
In this study
concordance between the ID results of Vitek-2
compact system and manual methods for all the
isolates of Enterobacter cloacae, Pseudomonas
aeruginosa, Enterococcus faecalis and Enterococcus
faecium was seen. However
discordant results majorly were seen for Klebsiella
species, Citrobacter species,
CONS and Staphylococcus aureus by
Vitek-2 compact system. Furthermore, appropriate
incubation conditions and duration should be
specified for organisms having slow metabolic
rates and late lactose fermenters like S.
hominis that are prone to be misidentified
by the Vitek-2 system. The
identification part of vitek-2 compact system has
flaws which need to be worked upon especially for
the organisms which cause serious life-threatening
infections (Salmonella spp, E. coli). The
treatment modality of the patients changes if the
identification of the organism is compromised
because separate group of antibiotics need to be
employed for treatment. The organisms having slow
metabolic rates are prone to errors by the Vitek-2
compact system. Incorporation of additional
biochemical tests like indole into the Vitek-2
cards can improve the identification and resolve
errors where Indole aids in identification (Salmonella
spp, E. coli, K. pneumonia, K. oxytoca).
Limitation
of the study: The small number of
samples collected may not adequately represent the
diversity of pathogens and resistance patterns
encountered in neonatal septicemia. While
identification is important, the accuracy and
reliability of antimicrobial susceptibility
testing (AST) results from the VITEK-2 system
should also be evaluated. Errors in AST can lead
to inappropriate antibiotic therapy choices. To
address these limitations, future studies could
focus on increasing sample size to improve the
robustness and generalizability of findings.
Additionally, incorporating AST evaluation using
the VITEK-2 system would enhance understanding of
its performance in antimicrobial susceptibility
testing, thereby providing more comprehensive data
for clinical decision-making.
Conclusion
In our study overall
concordance is 88.2% and discordance is 11.8%. It
is necessary to implement identification methods
that facilitate access to fast and reliable
results, but at the same time, help to optimize
the economic resources once those are implemented
in the daily routine. By implementing
identification methods that balance speed,
reliability, and economic efficiency, one can
enhance diagnostic capabilities in clinical
microbiology. This approach not only improves
patient care through timely and accurate treatment
decisions but also optimizes resource allocation
in the laboratory setting.
References
- Durand S. Executive summary—the globalization
of infectious disease. 2000. Available at http://www.prcdc.org/files/Infectious
Disease.pdf.
- Koneman E, Allen S. Koneman. Diagnostico
Microbiologico/ Microbiological diagnosis: Texto
Y Atlas En Color/ Text and Color Atlas. Ed.
Médica Panamericana; 2008.
- Deku JG, Dakorah MP, Lokpo SY, Orish VN,
Ussher FA, Kpene GE, et al. The Epidemiology of
Bloodstream Infections and Antimicrobial
Susceptibility Patterns: A Nine-Year
Retrospective Study at St. Dominic Hospital.
J Trop Med. 2019;p. 1–10.
- Alizadeh AM, Movahed RK, Mohammadnia M.
Comparative evaluation of conventional and
Bactec methods for detection of bacterial
infection. Tanaffos. 2016;15(2):112–6.
- Murray P, Baron E, Pfaller M, Tenover F,
Yolken R, editors. Manual of clinical
microbiology, 7th ed. American Society for
Microbiology, Washington, D.C; 1999.
- Duggal S, Gaind R, Tandon N, Deb M, Chugh T.
Comparison of an Automated System with
Conventional Identification and Antimicrobial
Susceptibility Testing. ISRN Microbiology.
2012;2012:1-4.
- Jossart M, Courcol R. Evaluation of an
automated system for identification of
Enterobacteriaceae and nonfermenting bacilli. European
Journal of Clinical Microbiology and
Infectious Diseases. 1999;
18(12):902-907.
- Paim TGS, Cantarelli VV, D’Azevedo PA.
Performance of the VITEK 2 system software
version 5.03 in the bacterial identification and
antimicrobial susceptibility test: evaluation
study of clinical and reference strains of
Gram-positive cocci. Rev Soc Bras Med Trop.
2013;47(3):377–81.
- Kuper K, Boles D, Mohr J, Wanger A.
Antimicrobial susceptibility testing: A primer
for clinicians. Pharmacotherapy. 2009;29(11):1326-1343.
- Doern GV, Vautour R, Gaudet M, Levy B.
Clinical impact of rapid in vitro susceptibility
and bacterial identification. J. Clin.
Microbiol. 1994;32:1757-1762.
- Cunha MLRS, Sinzato YK, Silveira LVA.
Comparison of methods for the identification of
coagulase negative staphylococci. Mem Inst
Oswaldo Cruz. 2004;99(8):855–60.
- Donay J, Mathieu D, Fernandes P, Pregermain C,
Bruel P, Wargnier A, et al. Evaluation of the
automated phoenix system for potential routine
use in the Clinical Microbiology Laboratory. Journal
of Clinical Microbiology. 2004;42(4):1542
1546.
- Nadheema H, Khetam HR, Jumaah DH. Frequency of
extended spectrum beta lactamase producing gram
negative bacteria isolated from blood cultures
at children hospital in Baghdad. IJSR -
International Journal of Scientific Research.
2015;4(1):10-12.
- Akgun S, Sayiner HS. Comparison of Rapid and
Routine Methods of Identification and Antibiotic
Susceptibility Testing of Microorganisms from
Blood Culture Bottles. Pol J Microbiol.
2020;69(2):1-12.
- Lupetti A, Barnini S, Castagna B, Capria AL,
Nibbering PH. Rapid identification and
antimicrobial susceptibility profiling of
Gram-positive cocci in blood cultures with the
Vitek 2 system. Eur J Clin Microbiol Infect
Dis. 2010;29(1):89-95.
- Chen Y, Porter V, Mubareka S, Kotowich L,
Simor AE. Rapid identification of bacteria
directly from positive blood cultures by use of
a serum separator tube, smudge plate
preparation, and matrix-assisted laser
desorption ionization–time of flight mass
spectrometry. J Clin Microbiol.
2015;53(10):3349–3352.
- Jagdish L, Naik TB, Gupta RK, Jais M. Etiology
of blood culture from septicemia cases and their
antibiotic susceptibility pattern at a tertiary
care hospital. Indian J Microbiol Res. 2016;3(4):435-9.
|



















