|
Introduction
Sepsis
is a life-threatening organ disfunction due to
dysregulated host response to infection (1). As
per World Health Organization (WHO), in 2017, 48.9
million cases and 11 million sepsis-related deaths
were reported worldwide, which accounted for
almost 20% of total global mortality (2). The
burden of the disease is evidently varied across
the geographical regions, approximately 85% of
these sepsis cases and sepsis-related deaths
occurred in low- and middle-income countries (2).
The striking increase in sepsis cases is largely
attributed to health care-associated infections.
Escalating prevalence of multi drug resistance
bacteria serves to exacerbate the situation, much
like adding fuel to the fire making it extremely
challenging to manage such life threatening
situations (3). Prompt intervention become vital
in saving the life of the patient, the Surviving
Sepsis Campaign guidelines also recommends early
initiation of antimicrobial therapy within an hour
of the detection of septic shock (4). Delay in the
administration of the appropriate antimicrobial
treatment is associated with higher mortality
rates and adverse consequences. Reducing the time
to identification and susceptibility testing is an
essential prerequisite to speed up targeted
antimicrobial therapy particularly in critically
ill patients with bloodstream infections (BSIs)
(5). Rapid and reliable techniques for isolation,
detection and antimicrobial susceptibility testing
(AST) of the causative pathogen is need of the
hour to adapt clinical intervention as fast as
possible.
European Committee
on Antimicrobial Susceptibility Testing (EUCAST)
has defined a methodology for rapid antimicrobial
susceptibility testing (RAST) directly from
positive blood culture bottles by disk diffusion
method with breakpoints for short incubations of
4hr, 6 hr and 8 hr. This method holds advantage of
quicker turn-around time over conventional AST
methods which usually requires a 16-20 hour of
incubation for pure growth followed by 16-20 hour
for AST (6,7). Clinical and Laboratory Standards
Institute (CLSI) also included direct
antimicrobial susceptibility testing (DAST) by
disk diffusion method AST using short incubation
times from the positive blood culture broth for
gram negative bacteria (GNB) (Enterobacterales
and Pseudomonas aeruginosa) (8). The
implementation of these novel approaches to
reduces turn-around time (TAT) for AST can provide
a reliable tool to improve clinical management of
sepsis patients. These guidelines (EUCAST and
CLSI) provide zone diameters for a limited number
of bacteria while for others the zone diameters
are yet to be established (6,8). Performed
according to guidelines, these methods are both
affordable, reliable and can be rapidly adapted to
new antimicrobials making them particularly
valuable in settings with limited resources where
advanced AST systems are not accessible (9-11).
This study attempted
to estimate/evaluate the performance of RAST
breakpoints directly from flagged BacT/Alert blood
culture bottles in clinical samples of suspected
sepsis patients. We also evaluated for the
presence of phenotypic drug resistance at the 4hr,
6hr and 8 hours of incubation.
Material & Methods
A descriptive cross-sectional study conducted at
the bacteriology laboratory of University College
of Medical Sciences, a tertiary care hospital in
Delhi, India, over a period of two months
(November – December, 2023). The non-duplicate
clinical samples of blood received in our
laboratory for routine culture were included in
the study.
Procedure
The RAST was
performed directly from the clinical samples for
blood cultures in parallel with the routine
testing. Blood cultures were routinely incubated
in BacT/Alert 3D system. The gram stain from the
flagged blood culture bottles were prepared to
identify the mono-bacterial growth and eliminate
poly-bacterial and mixed species growth. Within
the 0-18 hours of the flag time, the RAST was
performed as per EUCAST guidelines on flagged
blood cultures showing mono-bacterial growth.
About 125 to 150μL of undiluted blood culture
broth was lawned onto Mueller- Hinton agar and
antibiotic disks were placed evenly spaced across
the MHA and plates were incubated at 35–37 °C
under ambient condition. The antimicrobial disks
used for RAST were Ceftazidime (10 µg), Cefotaxime
(5µg), Piperacillin-Tazobactam (30/6 µg), Imipenem
(10µg), Meropenem (10 µg), Ciprofloxacin (5 µg),
Levofloxacin (5µg), Gentamicin (10 µg), Amikacin
(30 µg), tobramycin (10 µg), and
Amoxicillin-clavulanic acid (20/10 µg), for
Gram-negative bacilli (GNB). For gram-positive
cocci (GPC), Cefoxitin (30 µg), Norfloxacin (10
µg), Amikacin (30µg), Gentamicin (10µg and 30 µg),
Tobramycin (10 µg), Clindamycin (2 µg), disks were
used.
The isolates
identified as Escherichia coli, Klebsiella
pneumoniae, Pseudomonas aeruginosa,
Acinetobacter baumannii, Enterococcus faecalis,
Enterococcus faecium Staphylococcus aureus,
and Streptococcus pneumoniae were
included in the study. The identification of the
isolates was confirmed by Vitek-2 compact system.
Isolates other than as mentioned or displaying
mixed growth were excluded from the study. In
parallel with the RAST, routine testing was
performed and AST was done as per CLSI guidelines.
The inhibition zones
were read at 4hr, 6hr, 8hr and 16-20 hour of
incubation for Escherichia coli, Klebsiella
pneumoniae and Staphylococcus aureus.
Zones were read at 4hr, 6hr and 8hour of
incubation for Acinetobacter baumannii,
Enterococcus faecalis and Enterococcus
faecium and at 6hr and 8 hours16-20 hours
for Pseudomonas aeruginosa isolates. The
area of technical uncertainty (ATU) is the
zone-range where zone edges were not clearly
visible or there is an overlap of breakpoints of
susceptible and resistant isolates and hence it
cannot be determined whether the isolates are
susceptible or not.
Phenotypic
drug resistance
In addition to RAST,
phenotypic drug resistance mechanism for MRSA
(Methicillin-Resistant Staphylococcus aureus),
ICR (Inducible Clindamycin Resistance), ESBL
(Extended-Spectrum Beta-Lactamase) and CRE
(Carbapenem Resistance Enterobacterales)
were observed. For ESBL, double-disk synergy test
by using Ceftazidime (30 µg) and Ceftazidime plus
clavulanic acid (30/10 µg) disks were used. For
CRE, carbaNP (RAPIDEC® CARBA NP test) test was
used. In Staphylococcus aureus isolates,
MRSA was detected by using cefoxitin (30 µg) disks
while performing RAST and for ICR, D-test (zone of
inhibition around clindamycin appears as a letter
"D") was observed by placing Clindamycin (2 µg)
and Erythromycin (15 µg) disks adjacent to each
other.
For quality control,
Escherichia coli ATCC 25922, Pseudomonas
aeruginosa ATCC 27853 and Staphylococcus
aureus ATCC 25921 were used with each
batch as per the EUCAST guidelines. A purity plate
was put to observe contamination during the
procedure.
Statistical
analysis
For statistical
analysis, Microsoft SPSS version 2.0 was used. The
results from the RAST method were compared using
CLSI M100 guidelines as the reference method. The
categorical agreement (CA), very major errors
(VME), and major errors (ME) were determined. The
CA was defined as agreement in the interpretation
of the RAST and the reference method. VME (false
susceptibility) determines the percentage of false
susceptible results by RAST method as compared to
the reference method whereas ME (false resistance)
determines the percentage of false resistant
results by RAST methods as compared to the
reference method. As per CLSI recommendations, a
new system can be acceptable when it meets the
standards as follows: CA ≥ 90%, VME ≤ 1.5%, and ME
≤ 3% (CLSI, 2021). The ATU were not included in
the calculation of CA, VME and ME (12,13).
Results
Distribution
of isolates
Over a period of two
months (November – December, 2023), among flagged
BacT/Alert blood culture bottles, 42 showed
mono-bacterial isolates that qualified for further
testing by RAST method. Among these 42 clinical
isolates, 22 (52.4%) were gram positive cocci
(GPC) including Staphylococcus aureus 20
(47.6%) and Enterococcus fecalis 2
(4.8%) and 20 (47.6%) were GNB including Klebsiella
pneumoniae 10 (23.8%), Escherichia
coli 7 (16.7%) and Acinetobacter
baumannii 3 (7.1%).
A total of 42
clinical isolates, and 6 to 10 antimicrobials
tested per isolate resulted in overall 1259
inhibition zone diameters which were read at 4hr,
6hr, 8hr and 16-20 hour. The results of RAST are
summarized in Table 1.
|
Table 1: Readings of RAST
method of Staphylococcus aureus,
Klebsiella pneumoniae, Escherichia
coli and isolates at 4,6,8 and
16-20 hour as per EUCAST guidelines and Acinetobacter
baumannii at at 4,6 and 8 hour as
per EUCAST guidelines and at 16-20 hour as
per CLSI guidelines.
|
|
S. aureus (n= 20)
|
4 Hour
|
6 Hour
|
8 Hour
|
16-20 Hour
|
|
S
|
ATU
|
R
|
S
|
ATU
|
R
|
S
|
ATU
|
R
|
S
|
ATU
|
R
|
|
Cefoxitin
|
5
|
8
|
7
|
4
|
1
|
15
|
4
|
0
|
16
|
3
|
0
|
17
|
|
Norfloxacin
|
4
|
6
|
10
|
4
|
0
|
16
|
4
|
0
|
16
|
4
|
0
|
16
|
|
Amikacin
|
14
|
4
|
2
|
18
|
0
|
2
|
18
|
0
|
2
|
18
|
1
|
1
|
|
Gentamicin
|
9
|
10
|
1
|
11
|
4
|
5
|
13
|
2
|
5
|
13
|
2
|
5
|
|
Tobramycin
|
8
|
8
|
4
|
10
|
4
|
6
|
12
|
3
|
5
|
12
|
2
|
6
|
|
Clindamycin
|
10
|
7
|
3
|
15
|
1
|
4
|
15
|
0
|
5
|
15
|
0
|
5
|
|
K. pneumoniae (n=10)
|
4 hour
|
6 hour
|
8 hour
|
16-20 hour
|
|
Amoxicillin-clavulanic acid
|
0
|
6
|
4
|
1
|
3
|
6
|
1
|
2
|
7
|
0
|
0
|
10
|
|
Cefotaxime
|
1
|
1
|
8
|
2
|
0
|
8
|
2
|
0
|
8
|
2
|
0
|
8
|
|
Ceftazidime
|
1
|
3
|
6
|
0
|
2
|
8
|
1
|
1
|
8
|
0
|
2
|
8
|
|
Imipenem
|
6
|
2
|
2
|
5
|
3
|
2
|
4
|
3
|
3
|
7
|
1
|
2
|
|
Meropenem
|
2
|
2
|
6
|
1
|
1
|
8
|
1
|
1
|
8
|
1
|
1
|
8
|
|
Ciprofloxacin
|
1
|
2
|
7
|
2
|
0
|
8
|
2
|
0
|
8
|
2
|
1
|
7
|
|
Levofloxacin
|
0
|
2
|
8
|
1
|
1
|
8
|
1
|
1
|
8
|
1
|
1
|
8
|
|
Amikacin
|
3
|
2
|
5
|
4
|
0
|
6
|
4
|
0
|
6
|
4
|
0
|
6
|
|
Gentamicin
|
6
|
1
|
3
|
7
|
0
|
3
|
7
|
0
|
3
|
7
|
0
|
3
|
|
Tobramycin
|
6
|
2
|
2
|
7
|
0
|
3
|
7
|
0
|
3
|
7
|
0
|
3
|
|
E. coli (n= 7)
|
4 HOUR
|
6 HOUR
|
8 HOUR
|
16-20 HOUR
|
|
Amoxicillin-clavulanic acid
|
4
|
1
|
2
|
5
|
0
|
2
|
3
|
0
|
4
|
3
|
1
|
3
|
|
Cefotaxime
|
4
|
1
|
2
|
4
|
1
|
2
|
4
|
1
|
2
|
5
|
0
|
2
|
|
Ceftazidime
|
1
|
4
|
2
|
4
|
0
|
3
|
3
|
1
|
3
|
2
|
2
|
3
|
|
Imipenem
|
5
|
1
|
1
|
6
|
1
|
0
|
6
|
1
|
0
|
6
|
1
|
0
|
|
Meropenem
|
5
|
2
|
0
|
5
|
1
|
1
|
5
|
0
|
2
|
5
|
0
|
2
|
|
Ciprofloxacin
|
0
|
2
|
5
|
2
|
0
|
5
|
2
|
0
|
5
|
2
|
0
|
5
|
|
Levofloxacin
|
0
|
3
|
4
|
1
|
2
|
4
|
2
|
1
|
4
|
2
|
1
|
4
|
|
Amikacin
|
3
|
3
|
1
|
5
|
2
|
0
|
4
|
1
|
2
|
7
|
0
|
0
|
|
Gentamicin
|
5
|
1
|
1
|
6
|
0
|
1
|
6
|
0
|
1
|
6
|
0
|
1
|
|
Tobramycin
|
6
|
1
|
0
|
7
|
0
|
0
|
7
|
0
|
0
|
7
|
0
|
0
|
|
A.baumannii (n= 3)
|
4 HOUR
|
6 HOUR
|
8 HOUR
|
CLSI (Reference Method)
|
|
Imipenem
|
1
|
0
|
2
|
1
|
0
|
2
|
1
|
0
|
2
|
0
|
0
|
3
|
|
Meropenem
|
1
|
0
|
2
|
1
|
0
|
2
|
1
|
0
|
2
|
1
|
0
|
2
|
|
Ciprofloxacin
|
0
|
1
|
2
|
0
|
1
|
2
|
0
|
1
|
2
|
0
|
0
|
3
|
|
Levofloxacin
|
0
|
0
|
3
|
0
|
0
|
3
|
0
|
0
|
3
|
0
|
0
|
3
|
|
Amikacin
|
0
|
1
|
2
|
0
|
1
|
2
|
0
|
0
|
3
|
0
|
0
|
3
|
|
Gentamicin
|
1
|
0
|
2
|
0
|
0
|
3
|
0
|
0
|
3
|
0
|
0
|
3
|
|
Tobramycin
|
0
|
0
|
3
|
0
|
0
|
3
|
0
|
0
|
3
|
0
|
0
|
3
|
|
ATU: Area of technical
uncertainty; S: Susceptible; R: Resistant;
CLSI: Clinical and Laboratory Standards
Institute, RAST: Rapid antimicrobial
susceptibility testing
|
At 4 hours of
incubation, a thin but noticeable growth was
observed. However, by 6 hour to 8 hours, the
growth became clear and comparable to that of
16-20 hour of incubation. The ATU, which cannot be
determined whether the isolates are susceptible or
not were also noted. The percentage of results in
the ATU was more observed in early readings,
especially at 4 hour incubation. The percentage of
ATU at 4 hour was more observed in GPC (32%) as
compared to GNB (23%). However, their occurrence
significantly reduced at 6-hour incubation (8% in
GPC and 10% in GNB) and subsequent incubation
periods. Figure 1 illustrates the percentage of
ATU observed at 4hr, 6hr, 8hr and 16-20 hour of
incubation in GPC and GNB.
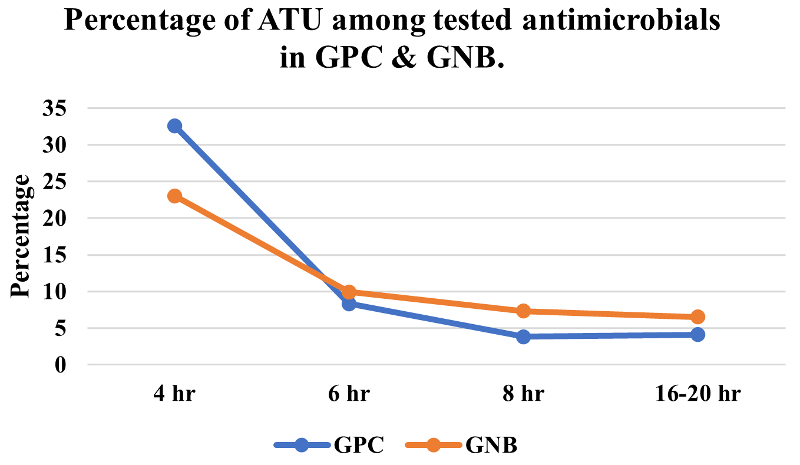
|
| Figure
1: Percentage of ATU among tested
antimicrobials in GPC and GNB at the time
of reading. [ATU: Area of Technical
Uncertainty, GPC: Gram-positive cocci,
GNB: Gram negative bacilli. |
Rapid
antimicrobial susceptibility testing in
Gram-positive cocci.
Among the S.
aureus isolates, none of the tested
antimicrobial presented with > 90% of
Categorical Agreement (CA) for any tested
antimicrobial at 4 hour of incubation. However the
highest CA (100%) was presented by Clindamycin 6
hour incubation and 90% at subsequent incubation
with no VME and ME. Cefoxitin, on the other hand,
achieved the CA >90% only after 8 hours of
incubation. There was no VME observed at 4,6,8
hour of incubation for any Cefoxitin, Gentamicin
and Clindamycin. However, up to 75% ME was
observed at 8 hour of incubation for Cefoxitin.
For Norfloxacin, the VME of 20% was observed at 4
hour incubation however no VME were observed at 6,
8 hour. Due to the absence of zone diameter for
Amikacin, and Tobramycin for Staphylococcus
aureus as per CLSI M100 guidelines,
calculations for categorical agreement, very major
errors, and major errors were not possible.
Among E.
fecalis, both the isolates exhibited zone
diameter within susceptibility category for
Linezolid and susceptible increased exposure (Si)
category for Imipenem at 4hr, 6hr and 8 hour of
incubation. The zone diameters for Vancomycin were
in ATU category and were not resistance for both
the isolates at 4hr, 6hr and 8 hour of incubation.
Additionally, for high level aminoglycoside
resistance (HLAR) testing, one isolate was
positive and one was negative for HLAR by using
Gentamicin (30ug) disk at 4hr, 6hr and 8 hour of
incubation. On comparing the RAST results with the
reference method, both the isolates were
susceptible for Imipenem, Vancomycin, Linezolid as
well as to high level Gentamicin (120ug) when AST
was performed as per CLSI guidelines.(Table 2)
|
Table 2: The comparison of
results of RAST to the reference method
(as per CLSI guidelines).
|
|
CLSI (Reference method)
|
4 Hour
|
6 Hour
|
8 Hour
|
16-18 Hour
|
|
S. aureus (20)
|
S
|
R
|
CA%
|
VME%
|
ME%
|
CA%
|
VME%
|
ME%
|
CA%
|
VME%
|
ME%
|
CA%
|
VME%
|
ME%
|
|
Cefoxitin
|
16
|
4
|
61.5
|
0
|
18.8
|
89.4
|
0
|
68.8
|
90
|
0
|
75.0
|
95
|
0
|
81.3
|
|
Norfloxacin
|
5
|
15
|
70.5
|
20
|
25
|
95
|
0
|
25
|
95
|
0
|
25
|
95
|
0
|
25
|
|
Gentamicin
|
16
|
4
|
50
|
0
|
37.5
|
63
|
0
|
6.3
|
73.6
|
0
|
6.3
|
75
|
0
|
6.3
|
|
Clindamycin
|
15
|
5
|
63.1
|
0
|
13.3
|
100
|
0
|
0
|
90
|
0
|
0
|
90
|
0
|
0
|
|
Enterobacteriales (17)
|
|
Amoxicillin-clavulanic acid
|
3
|
14
|
90
|
7.1
|
0
|
78.57
|
21.4
|
0
|
93.3
|
7.1
|
0
|
100
|
0
|
0
|
|
Cefotaxime
|
0
|
17
|
66.6
|
29.4
|
0
|
66.60
|
35.3
|
0
|
62.5
|
35.3
|
0
|
62.5
|
41.2
|
0
|
|
Ceftazidime
|
3
|
14
|
58.3
|
0
|
0
|
75
|
7.1
|
0
|
88.23
|
7.1
|
0
|
92.3
|
0
|
0
|
|
Imipenem
|
7
|
10
|
42.1
|
40.0
|
0
|
69.2
|
0
|
0
|
64.28
|
30.0
|
0
|
61.53
|
60
|
0
|
|
Meropenem
|
6
|
11
|
66.6
|
9.1
|
0
|
100
|
0
|
0
|
100
|
0
|
0
|
100
|
0
|
0
|
|
Ciprofloxacin
|
4
|
13
|
92.3
|
23.1
|
0
|
88.8
|
0
|
0
|
88.2
|
0
|
0
|
87.5
|
0
|
0
|
|
Levofloxacin
|
7
|
10
|
73.3
|
0
|
28.6
|
86.6
|
0
|
28.6
|
82.3
|
0
|
28.6
|
93.3
|
0
|
28.6
|
|
Amikacin
|
5
|
12
|
61.5
|
8.3
|
0
|
57.1
|
33.3
|
0
|
68.75
|
25.0
|
0
|
52.94
|
50
|
0
|
|
Gentamicin
|
12
|
5
|
82.3
|
0
|
0
|
94.11
|
20
|
0
|
94.11
|
0
|
0
|
94.11
|
20
|
0
|
|
Tobramycin
|
5
|
12
|
31.2
|
58.3
|
0
|
42.1
|
0
|
0
|
47
|
75.0
|
0
|
47
|
75
|
0
|
|
CA : Categorical
agreement, VME: Very major error, ME:
Major error, AST : Antimicrobial
susceptibility testing, RAST: Rapid
antimicrobial susceptibility testing, CLSI
: Clinical and Laboratory Standards
Institute, and EUCAST: European Committee
on Antimicrobial Susceptibility Testing.
|
Rapid
antimicrobial susceptibility testing in Gram-
negative bacilli
K. pneumoniae
was the most common isolate among GNB, followed by
E.coli. Majority of antimicrobials
(except Amoxicillin-clavulanate and Ciprofloxacin)
had CA of < 90% at 4 hour incubation for Enterobacteriales.
The VME observed at 4 hour incubation was high
(>1.5%) for most of the antimicrobials (Except
for Ceftazidime, Levofloxacin and Gentamicin).
While the ME was high for Levofloxacin (28.6%),
and rest of the antimicrobials showed no ME (0%).
At 6 hour incubation only 2 antimicrobials
(Meropenem and Gentamicin) had CA of > 90%. No
VME was observed for 5 antimicrobials (Imipenem,
Meropenem, Ciprofloxacin, Levofloxacin and
Tobramycin) and high (>1.5%) VME was observed
for rest of the antimicrobials. No ME was observed
at 6 hour incubation for most of the
antimicrobials (except Levofloxacin). No much
variation in results of CA,VME and ME was observed
at 8 hour incubation as compared to the results at
6 hour incubation.
Among gram negative
isolates, 3 were Acinetobacter baumannii,
on comparing RAST results with reference method,
Levofloxacin, Meropenem and Tobramycin achieved
same result at 4, 6 and 8 hour. However Gentamicin
and Amikacin achieved the same result at 6 hour
and 8 hour incubation respectively.
Antimicrobial
resistance
Antimicrobial
resistance (MRSA, ICR, ESBL and CRE) was observed.
At 6 hour incubation, 32% of AMR was recorded and
remaining 68% at 8 hour of incubation. The
percentage of antimicrobial resistance detected is
shown in Figure 2.

|
| Figure
2: Distribution of antimicrobial
resistance among tested isolates in the
study group. [MRSA: Methicillin Resistant
Staphylococcus aureus; ICR: Inducible
clindamycin resistance; ESBL: Extended
Spectrum Beta-Lactamase; CRE:
carbapenem-resistant Enterobacterales.]
|
|
| Figure 3: RAST of gram
negative bacteria at 4 hour incubation
(a); at 6 hour incubation (b); at 8 hour
incubation (c) [RAST: Rapid antimicrobial
susceptibility testing] |
Discussion
The foundation of
sepsis management relies on promptly administering
suitable and efficient antimicrobial therapy.
However, the prevalence of MDR pathogens
complicates the empirical antibiotic treatment
choices, increases the risk of inappropriate
treatments. The conventional methods usually take
2-3 days for identification and susceptibility
testing of the causative pathogens from positive
blood cultures. The utilization of RAST method
reduces the duration of susceptibility testing,
thereby expediting the implementation of targeted
antimicrobial therapy. In this study we observed
and compared the results of RAST by EUCAST with
reference method (CLSI) along with phenotypic
antimicrobial resistance.
The EUCAST
guidelines defines ATU as zone range where the
edges of zones are not distinctly visible or where
breakpoints of susceptible and resistant isolates
overlap, making it challenging to determine the
susceptibility of the isolates. In our study, the
ATU percentage was high at 4 hour incubation which
reduces drastically at subsequent incubation
period. The similar findings were observed in
another study by Cherkaoui et al (2023)
(13). They observed a high rate of ATU for E.
coli, Klebsiella spp., A.
baumannii and S. aureus isolates
was observed at 4 h, but this rate declined over
time. In contrast to Cherkaoui et al, we
observed a higher occurrence of ATU in GPC as
compared to GNB after 4 hour incubation.
In our study, for S.aureus,
the CA of < 90 % and no VME was observed at 4
hour of incubation with high ME for most of the
tested antimicrobial. The CA remained same for the
GNBs however the high VME, and no ME was observed
in Enterobacteriales for most of the
antimicrobials tested. In another study by Soo et
al (2019), They compared the RAST results
of 194 gram negative isolates against the routine
method, Vitek 2. They observed high ME rates for
Piperacillin-tazobactam and ceftazidime in gram
negative isolates (E.coli and K.
pneumoniae) similar to our study. However
the VME rates were low at 4 hour incubation and CA
of > 90% was achieved in their which is not
achieved in our study (14,15).
In a study by Park et
al. (2023), for S. aureus, they
observed high CA,(100%) at 4, 6, and 8 hours.
There were no ME or VME at 4, 6, and 8 hours. All
categories were consistent between the two methods
when compared with the broth microdilution
(Sensitiser) results based on the CLSI criteria.
In our study discrepancy among the CA and ME were
observed in S. aureus, though the CA of
> 90% was achieved by most of the
antimicrobials at 8 and 16-18 hour (16).
At results of RAST
at 6 hour and 8 hour incubation is almost similar
with most of the antimicrobials. In a study by
Mancini et al, (2020) the overall better
separation of the susceptible and resistant
strains after 8 h incubation resulted in generated
a higher number of interpretable results as
compared to results after 6 hour incubation unlike
our study (17). This may be attributed to the
small sample size and the utilization of automated
techniques for inoculum preparation (such as flow
cytometry using the UF-4000 system) and streaking
to minimize human errors, aspects that were not
present in our study (17).
With the suggested
breakpoints of RAST, up to 32% of AMR (MRSA, ICR,
ESBL and CRE) can be detected phenotypically at 6
hour, and remaining 68% can be detected at 8 hour
of incubation. Similar results were found in study
by Jonson et al. (2020) (7).
RAST is a not a
complicated method to introduce in standard
clinical microbiology laboratories. It is
cost-effective, simple to execute, and provides
rapid results, particularly in settings with
limited resources where automated methods like
Vitek and Maldi-TOF are unavailable. Implementing
this method demands adaptation of work- flow to
reduce the human errors and enhance precision. The
reporting of the RAST results can be challenging
due to the increased likelihood of poor CA, VME
and ME during the initial hours of incubation
along with the ATU. Though the incorporation of
the ATU in the guidelines itself prevents
unavoidable variation from causing VME and ME to
some extent. We noted an overall increase in CA
and more favourable VME and ME for selected
antimicrobials. Hence , RAST can be used to for
selected antimicrobials not all, but caution is
advised, particularly regarding early readings,
especially the 4-hour reading.
Conclusion
The RAST method
offers rapid results, yet there are some
limitations in evaluating the applicability of
this method. Unlike CLSI, EUCAST-RAST method
cannot be performed for various strains and
antimicrobial agents. As of now, the RAST method
has only undergone validation for eight species
and is not applicable to species beyond this
validated set. In this study we observed that the
number of ATU was high after 4 hour incubation
making it unreliable for reporting. The
discrepancy among CA, VME and ME undermines the
reliability of the test for certain antimicrobials
at 6 and 8 hours of the test. Though in general,
the numbers of VME and ME decreased over time and
CA increases with subsequent incubations. The
early phenotypic detection of antimicrobial
resistance at 6-8 hours significantly aids in
initiating antimicrobial treatment. This study
indicates that RAST method can be used to for
certain antimicrobials not all, also the early
readings, especially the 4-h reading, using the
RAST breakpoints proposed by EUCAST cannot be used
clinical microbiology laboratories. The primary
lacunae of this study is its limited sample size.
Increased sample size is imperative to ensure more
precise results.
References
- Rudd KE, Johnson SC, Agesa KM, Shackelford KA,
Tsoi D, Kievlan DR, et al. Global, regional, and
national sepsis incidence and mortality,
1990-2017: analysis for the Global Burden of
Disease Study. Lancet.
2020;395(10219):200-11.
- World Health Organisation. Fact Sheet: Sepsis.
Available at https://www.who.int/news-room/fact-sheets/detail/sepsis.
2024.
- Iancu D, Moldovan I, Țilea B, Voidăzan S.
Evaluating Healthcare-Associated Infections in
Public Hospitals: A Cross-Sectional Study. Antibiotics.
2023 Dec 2;12(12):1693.
- Evans L, Rhodes A, Alhazzani W, Antonelli M,
Cooper-smith CM, French C et al. Surviving
sepsis campaign: international guidelines for
management of sepsis and septic shock 2021. Intensive
Care Med. 2021; 47(11), 1181-247.
- Marchaim D, Gottesman T, Schwartz O et al.
National multicenter study of predictors and
outcomes of bacteremia upon hospital admission
caused by Enterobacteriaceae producing
extended-spectrum b-lactamases. Antimicrob
Agents Chemother 2010; 54: 5099–104.
- EUCAST. Breakpoint Tables for Interpretation
of MICs and Zone Diameters, Version 8.1. 2018. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.1_Breakpoint_Tables.pdf.
- Jonasson E, Matuschek E, Kahlmeter G. The
EUCAST rapid disc diffusion method for
antimicrobial susceptibility testing directly
from positive blood culture bottles. J
Antimicrob Chemother 2020; 75: 968–78.
- CLSI. M100Ed33. Performance standards for
antimicrobial susceptibility testing: 33rd
informational supplement. Wayne, PA: Clinical
and Laboratory Standards Institute; 2023.
- Ahman J, Matuschek E, Kahlmeter G. The quality
of antimicrobial discs from nine
manufacturers—EUCAST evaluations in 2014 and
2017. Clin Microbiol Infect.
2019;25:346–52.
- Ahman J, Matuschek E, Kahlmeter G. EUCAST
evaluation of 21 brands of Mueller-Hinton
dehydrated media for disk diffusion testing. Clin
Microbiol Infect 2020;
doi:10.1016/j.cmi.2020.01.018.
- Brown D, Canton R, Dubreuil L et al.
Widespread implementation of EUCAST breakpoints
for antibacterial susceptibility testing in
Europe. Euro Surveill 2015; 20: 21008.
- CLSI. M100Ed31. Performance standards for
antimicrobial susceptibility testing: 31st
informational supplement. Wayne, PA: Clinical
and Laboratory Standards Institute; 2021.
- Cherkaoui A, Schorderet D, Azam N, Crudeli L,
Fernandez J, Renzi G, Fischer A, Schrenzel J.
Fully Automated EUCAST Rapid Antimicrobial
Susceptibility Testing (RAST) from Positive
Blood Cultures: Diagnostic Accuracy and
Implementation. Journal of Clinical
Microbiology. 2022 Oct
19;60(10):e00898-22.
- Soo YT, Waled SN, Ng S, Peh YH, Chew KL.
Evaluation of EUCAST rapid antimicrobial
susceptibility testing (RAST) directly from
blood culture bottles. European Journal of
Clinical Microbiology & Infectious
Diseases. 2020 May;39:993-8.
- Heather CS, Maley M. Automated direct
screening for resis- tance of gram-negative
blood cultures using the BD Kiestra Work Cell. Eur
J Clin Microbiol Infect Dis
2018;37(1):117–125.
- Park JM, Kwon M, Hong KH, Lee H, Yong D.
European Committee on Antimicrobial
Susceptibility Testing-Recommended Rapid
Antimicrobial Susceptibility Testing of
Escherichia coli, Klebsiella pneumoniae, and
Staphylococcus aureus From Positive Blood
Culture Bottles. Annals of Laboratory
Medicine. 2023 Sep 1;43(5):443-50.
- Mancini S, Bodendoerfer E, Kolensnik-Goldmann
N, Herren S, Röthlin K, Courvalin P, Böttger EC.
Evaluation of standardized automated rapid
antimicrobial susceptibility testing of
Enterobacterales-containing blood cultures: a
proof-of-principle study. Journal of
Antimicrobial Chemotherapy. 2020
Nov;75(11):3218-29.
|



















