|
Introduction
Antimicrobial
resistance is a top priority worldwide in
healthcare system. In 2017, WHO published a list
of global priority pathogens, including
carbapenem-resistant Enterobacterales,
carbapenem-resistant Pseudomonas aeruginosa,
and carbapenem-resistant-Acinetobacter
baumannii under the category of critical
organisms. (1,2) The available antibiotics have
either lost their ability to treat bacterial
infections or are going to lose very quickly. The
increasing prevalence of antimicrobial resistance
in Gram-negative bacteria is a matter of serious
concern, as the acquisition of antimicrobial
resistance in gram-negative bacteria is more rapid
than that in Gram-positive bacteria. (3-6)
The
β-lactam group of antibiotics is the most
prevalently prescribed antibiotic. The widespread
use of antibiotics in the community as well as in
hospital practices has induced selection pressure
toward resistant strains and favoured the
evolution of antimicrobial resistance.(7)
Gram-negative bacteria have inherent abilities to
become resistant to antimicrobials and also
facilitate other bacteria to become
drug-resistant.(8,9) Spontaneous random mutations
and the transfer of genes either horizontally or
vertically can evolve resistant strains.(10)
Enzymatic
drug inactivation of β-lactam antibiotics by
β-lactamases (e.g., AmpC, ESBLs, carbapenemases)
is the primary cause of resistance to β-lactam
antibiotics, especially in gram-negative
bacteria.(11) These β-lactamases differ
structurally as well as characteristically from
each other. Various widespread
β-lactamase-mediated resistance mechanisms can be
detected in the microbiology laboratory following
CLSI (Clinical and Laboratory Standards Institute)
guidelines and other standard references without
the involvement of any molecular biology
techniques and can help in the assessment of the
prevalence of β-lactamases in the region. During
the COVID-19 pandemic which lasted from December,
2019-21, there was a widespread use of
broad-spectrum antibiotics in COVID-19 patients,
which was considered responsible for further
escalation of antimicrobial resistance. (12,13)
We conducted this study to find out the local
prevalence of β-lactamases in Gram-negative
bacteria in our region.
Methods
This cross-sectional
study was conducted in a multi-specialty hospital
located in Central India from 01st January 2021 to
31st December 2023. In this study, all clinical
specimens submitted in microbiology department
from in-patient and outpatient departments of
hospital were included.
Preliminary
identification of organisms isolated from clinical
samples was performed by Gram staining, colonial
characteristics, and biochemical characterization
of the bacteria. (14) Confirmed identification was
done by automated system.
The antimicrobial
susceptibility of all 691 gram-negative isolates
to antimicrobials was determined by both the broth
microdilution method and manually by the Kirby
Bauer disk diffusion method. Selection of
antimicrobials and interpretation of MIC (minimum
inhibitory concentration) and zone diameter
results were checked in accordance with CLSI
guidelines. An additional colistin broth disk
elution test was performed by using colistin (10
μg) discs to evaluate the MIC of colistin against
the isolates. (15)
Analysis of
antimicrobial resistance due to β-lactamase
production in gram-negative bacteria was detected
by phenotypic methods. The basic β-lactamases
detected were ESBL, AmpC BL and
metallo-β-lactamase by standard manual methods
described in CLSI. The methods that we used were
simple and performed manually on a routine basis.
ESBL and AmpC
production in the gram-negative isolates was
checked by performing the double disc synergy test
recommended by CLSI. The antibiotic disks
containing cefotaxime (30 μg), cefotaxime
clavulanic acid (30/10 μg), ceftazidime(30 μg),
ceftazidime clavulanic acid (30/10 μg), cefoxitin
(30 μg), imipenem (10 μg), imipenem-EDTA (10/750),
and aztreonam (30 μg) were used for resistance
detection. Bacteria were classified as ESBL
positive when the zone of inhibition around the
third-generation cephalosporin and clavulanic
acid-containing combination disk was 5 mm higher
than the zone obtained by only the
third-generation cephalosporin disk.
Complete resistance
against β-lactams and β-lactamase inhibitor
combinations, cephalosporins up to third
generation and cefoxitin were used to predict a
high level of AmpC production. (20)
Phenotypic detection
of metallo β-lactamase production was checked by
using the imipenem EDTA disk method. An increase
in zone diameter around the Imipenem EDTA disk by
≥ 7 mm compared to the Imipenem -containing disk
confirms the presence of MBL in the isolate
tested. (17, 22, 23)
Statistically,
significance was considered at p value less than
0.05 by using Chi-square test. Data analyses were
conducted using the Excel software.
Result
In this study, we
included a total of 3710 clinical samples received
in the Microbiology Department for culture tests
between 2021-2023. Out of 3710 cultures, 991 (27%)
cultures were positive with significant
microscopic findings.
The percentage of
gram-negative bacilli was 65% of the total
positive samples, followed by gram-positive cocci
(32%) and fungi (2%). E. coli (41%) was
found to be the most prevalent pathogen followed
by Pseudomonas aeruginosa (15%), Salmonella
spp. (15%) and, Klebsiella pneumoniae
(11%). Our findings suggest that 74% of the total
gram-negative bacilli belong to the family Enterobacterales.
Here, we studied 285
cultures of E. coli isolated from
various specimens, such as blood, urine and pus.
Total of 77% were found to be associated with
urinary tract infections.
Klebsiella
was the third most predominant organism isolated,
that belongs to the family Enterobacterales.
We obtained 78 isolates identified as Klebsiella
pneumoniae, of which 54% were from urine
samples, 26% were from pus samples, 13% were from
respiratory samples, and the remaining 7% were
from blood cultures.
Pseudomonas
is a known pathogen associated with various
infections, especially nosocomial infections. In
our study, the prevalence of Pseudomonas
was 15% of the total Gram-negative isolates.
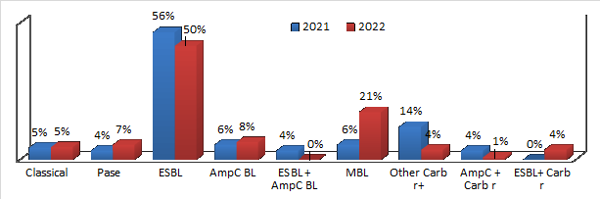
|
| Figure
1. Comparative analysis of antimicrobial
resistance in E. coli |
Antimicrobial
susceptibility testing and antimicrobial
resistance profiling of E. coli isolates
revealed that more than 85% of isolates are
resistant to the most often used third-generation
cephalosporin class of antibiotics.
The extended
spectrum β-lactamase resistance mechanism was the
leading antimicrobial resistance detected in 53%
of total E. coli isolates, followed by
metallo β-lactamases (14%). Overall, 28% of the
isolates were found to be resistant to even
carbapenems, which is an alarming situation. Our
study revealed a 6% prevalence of MBL in E.
coli in the year 2021, which has now
increased to 21% by 2022. By comparing the data
from 2021 and 2022, we found an overall increase
in antimicrobial resistance, showing a significant
15% increase in metallo β-lactamases (Figure 1).
The therapeutic choice suggested for the
infections caused by ESBL-producing E. coli
was found to be meropenem with susceptibility
(79%) and amikacin (91%). In the case of lower
urinary tract infections, fosfomycin (99%
susceptibility) and nitrofurantoin (87%
susceptibility) have emerged as alternatives to
β-lactam antibiotics.
Antimicrobial
susceptibility testing of Klebsiella
showed an overall increasing trend in
antimicrobial resistance. It is also evident from
the resistance trend that the highest rise in
resistance was found against amikacin. The
amikacin resistance was found to increase by 30%
with a statistically significant p-value of
0.02325 (at p<0.05). The current study revealed
72% of the clinical isolates to be resistant to
third-generation cephalosporins. In addition to
the drastic drop in cephalosporin susceptibility,
susceptibility to carbapenems has also decreased.
Comparative analysis of resistance levels in 2021
and 2022 revealed an increase in the prevalence of
MBL resistance by 33% which is statistically
significant with p-value of 0.009937.
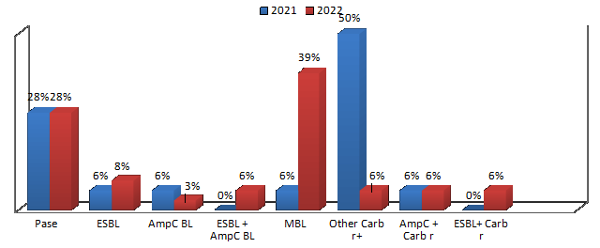
|
| Figure
2: Comparative analysis of antimicrobial
resistance in Klebsiella pneumoniae
|
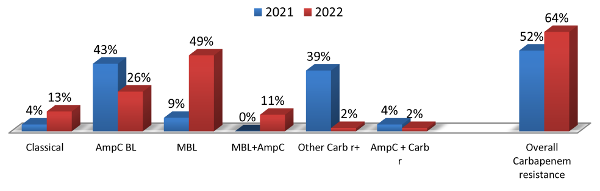
|
| Figure 3: Comparative
analysis of antimicrobial resistance in P.
aeruginosa |
Among the range of
drugs analysed in the current study, we observed
that 53% of the Pseudomonas aeruginosa
isolates were susceptible to piperacillin
tazobactam, 50% were susceptible to aztreonam, 49%
were susceptible to meropenem and 51% were
susceptible to gentamicin. The matter of major
concern in our study is that the prevalence of MBL
increased by 51% from 2021 to 2022 which is
statistically significant with p value of 0.009937
(Figure 3).
Discussion
Out of 3710 samples
received in microbiology laboratory between
2021-23, 990 (27%) were culture positive with 65%
being Gram-negative bacilli, 25% Gram-positive
cocci and 2% were fungus. Among Gram-negative
bacilli, 41% were E. coli being most
prevalent followed by Pseudomonas aeruginosa
(15%), Salmonella Typhi (15%) and Klebsiella
pneumoniae (11%). In the neighbouring
country of India, Nepal, a study was conducted
that showed nearly the same prevalence of
organisms viz. E. coli (46.7%), followed
by Klebsiella and Pseudomonas spp.
which is in concordance with our study. (16)
Our findings suggest
that 74% of the total gram-negative bacilli belong
to the family Enterobacterales which is
similar to study conducted by Erdem et al., where
75-95% of community-acquired lower urinary tract
infections are due to E. coli. (17)
Shreshtha et al. reported the presence of 53%
ESBL-producing E. coli in their study
conducted in Nepal which is in accordance with our
study, that revealed 53% of E. coli
isolates to be ESBL resistant. (16) Our findings
were consistent with a study stating that ESBL
genes such as CTX-M, TEM and SHV families are most
frequently found in Enterobacterales
with ESBL CTX-M type as the most prevalent
resistance phenotype. (16,18)
In our study, we
observed the rising trend of metallo β-lactamases
similar to other various studies that reported the
widespread presence of metallo β-lactamases in the
Indian subcontinent. Clinical isolates may exhibit
the presence of one or more carbapenem resistance
genes, such as MBL blaNDM-1, OXA-48, or KPC. (19)
Data published by Kamble D. from Pune in 2015
stated a 5% prevalence of MBL in E. coli.
(20) In case of E. coli mediated urinary
tract infections Fosfomycin and Nitrofurantoin are
found to be effective as non β-lactam options
available.
In our study, we
found that the susceptibility of Klebsiella
pneumoniae to amikacin in 2021 was 83%, and
in 2022 it was 53%. As per study by Kuti et al.,
in 2018 found 83.7% susceptibility of Klebsiella
pneumoniae to amikacin. (21) As per study
by Bora et al., in 2014 there was 21.08%
prevalence of MBL-producing Klebsiella
pneumoniae in different clinical samples.
(22) The results of our study have also put
forward an alarming situation for carbapenem
resistance. Total of 57% of the clinical isolates
in the current study were carbapenem resistant,
and 28% of these isolates exhibited MBL-mediated
resistance.
In our study, we
observed that 59% of the Pseudomonas isolates
were carbapenem resistant, and out of that 43%
were MBL producers. Among all the isolates of P.
aeruginosa 31% showed inducible AmpC
β-lactamase-mediated resistance. Pseudomonas
exhibits intrinsic resistance against most
commonly used antibiotics, resulting in fewer
available options to treat infections. Colistin
and polymyxin B are considered the most effective
antipseudomonal drugs. The colistin broth disk
elution (CBDE) method was used to confirm the
colistin MIC. According to Kuti et al.’s findings
in 2018, the susceptibility of Pseudomonas
aeruginosa to ciprofloxacin was 83.9%, that
to ceftazidime was 81.3%, and that to cefepime was
80.4%. When compared to our findings, the data
from Kuti et al. indicated a significant decrease
in susceptibility to ciprofloxacin, ceftazidime,
and cefepime of 29%, 42%, and 52%, respectively,
for Pseudomonas aeruginosa. The same
research reported interesting findings on amikacin
resistance. Amikacin susceptibility was studied in
carbapenem-resistant and carbapenem-susceptible
strains. A total of 91.1% carbapenem-sensitive
strains were found to be susceptible to amikacin,
whereas only 80.9% carbapenem-resistant strains
were susceptible to amikacin. (21) We found 53% Pseudomonas
aeruginosa susceptible to amikacin.
The current study is
based on phenotypic expression of β-lactamases in
pathogens. There might be many other β-lactamases
exhibited by the isolates that might be
unexpressed, which can be further detected with
the help of molecular methods. This study was
conducted to learn more about the microorganisms
that possess potentially widespread antimicrobial
resistance mechanisms. The findings of the study
will help in preparing the region-specific
empirical antibiotic policy, which in turn will
help in limiting the injudicious use of
antibiotics.
The study findings
revealed that the three most common gram-negative
clinical isolates E. coli, K. pneumoniae and
P. aeruginosa, have a significantly higher
incidence of MBL. The ability of Pseudomonas to
develop resistance against available antibiotics
by following various mechanisms makes it a
dreadful pathogen. The increasing carbapenem
resistance in Pseudomonas may develop following
chromosomal mutations that decrease permeability
while also causing the overexpression of efflux
pumps and the production of β-lactamases. (23) The
rise in MBL points towards excessive use of
carbapenems in our healthcare, leading to
selection pressure as well as rapid transmission
of resistance across patients due to inefficient
infection control practices.
In such scenario
carbapenems have become an inefficient option and
thus it becomes difficult to treat infections
caused by these resistance-carrying pathogens. Use
of carbapenems as empiric therapy should strongly
discouraged with the help of effective antibiotic
stewardship programmes, and antibiotics
particularly higher ones should be prescribed only
with undeniable bacterial infections in which case
the empiric antibiotic therapy should be
immediately switched to targeted therapy based on
the results of culture and sensitivity testing.
There is also a need to implement diagnostic
stewardship practices in healthcare, so as to
reduce the incidence of inappropriate diagnosis of
infections resulting in irrational use of
broad-spectrum antibiotics.
Conflicting Interests:
The authors declared no
potential conflicts of interest.
Funding sources: None
Acknowledgements
The authors would
like to thank the management and colleagues of
institution for providing us the necessary support
to conduct the study. We would like to thank
Parvati Birla and Murli Patel for the technical
support.
References
- WHO. Global Antimicrobial Resistance and Use
Surveillance System (GLASS) Report. 2020.
- Asokan GV, Ramadhan T, Ahmed E et al. WHO
global priority pathogens list: A bibliometric
analysis of Medline-Pubmed for knowledge
mobilization to infection prevention and control
practices in Bahrain. Oman Med J. 2019;34(3):184–93.
- Exner M, Bhattacharya S, Christiansen B.
Antibiotic resistance: What is so special about
multidrug-resistant Gram-negative bacteria?
GMS Hyg Infect Control. 2017 Apr
10;12:Doc05.
- Miller SI. Antibiotic resistance and
regulation of the Gram-negative bacterial outer
membrane barrier by host innate immune
molecules. mBio. 2016 Sep 1;7(5).
- Gupta V, Datta P. Next-generation strategy for
treating drug resistant bacteria: Antibiotic
hybrids. Indian Journal of Medical Research.
2019;149:97–106.
- Breijyeh Z, Jubeh B, Karaman R. Resistance of
gram-negative bacteria to current antibacterial
agents and approaches to resolve it. Molecules.
2020 Mar 16;25(6):1340.
- Kwapong AA, Soares S, Teo SP et al. Myristica
lowiana Phytochemicals as Inhibitor of Plasmid
Conjugation in Escherichia coli.
Evidence-based Complementary and Alternative
Medicine. 2020 Jun 19;2020:1604638.
Available at https://pmc.ncbi.nlm.nih.gov/articles/PMC7321511/
- Nordmann P, Poirel L, Walsh TR. et al. The
emerging NDM carbapenemases. Trends in
Microbiology. 2011 Dec;19(12):588-95.
- Munoz-Price LS, Poirel L, Bonomo RA, et al.
Clinical epidemiology of the global expansion of
Klebsiella pneumoniae carbapenemases. The
Lancet Infectious Diseases. 2013
Sep;13(9):785–796. Available at https://pmc.ncbi.nlm.nih.gov/articles/PMC4673667/
- Wellington EMH, Boxall ABA, Cross P, et al.
The role of the natural environment in the
emergence of antibiotic resistance in
Gram-negative bacteria. The Lancet
Infectious Diseases. 2013
Feb;13(2):155-65.
- Queenan AM, Bush K. Carbapenemases: The
versatile β-lactamases. Clinical
Microbiology Reviews. 2007
Jul;20(3):440-58. Available at https://pmc.ncbi.nlm.nih.gov/articles/PMC1932750/
- Malik SS, Mundra S. Increasing Consumption of
Antibiotics during the COVID-19 Pandemic:
Implications for Patient Health and Emerging
Anti-Microbial Resistance. Antibiotics. 2022
Dec 28;12(1):45. Available at https://pmc.ncbi.nlm.nih.gov/articles/PMC9855050/
- Clancy CJ, Buehrle DJ, Nguyen MH. PRO: The
COVID-19 pandemic will result in increased
antimicrobial resistance rates. JAC
Antimicrob Resist. 2020 Sep 1;2(3).
- Cheesbrough Monica. District Laboratory
Practice in Tropical Countries. Second, 2006.
Vol. 2. Cambridge University Press;
- CLSI. CLSI M100-ED33:2023 Performance
Standards for Antimicrobial Susceptibility
Testing, 33rd Edition. 2023.
- Shrestha A, Acharya J, Amatya J, et al.
Detection of Beta-Lactamases (ESBL and MBL)
Producing Gram-Negative Pathogens in National
Public Health Laboratory of Nepal. Int J
Microbiol. 2022 Oct 6:2022:5474388.
Available at https://pmc.ncbi.nlm.nih.gov/articles/PMC9560861/
- Erdem I, Kara Ali R, Ardic E, et al.
Community-acquired lower urinary tract
infections: Etiology, antimicrobial resistance,
and treatment results in female patients. J
Glob Infect Dis. 2018 Jul 1;10(3):129–32.
- Paterson DL, Bonomo RA. Extended-spectrum
β-lactamases: A clinical update. Clinical
Microbiology Reviews. 2005;18:657–86.
- Boyd SE, Livermore DM, Hooper DC, et al.
Metallo-β-lactamases: Structure, function,
epidemiology, treatment options, and the
development pipeline. Antimicrob Agents
Chemother. 2020 Sep 21;64(10):e00397-20.
Available at https://pmc.ncbi.nlm.nih.gov/articles/PMC7508574/
- Kamble D. Phenotypic detection of ESBL and MBL
in Gram-negative bacilli isolated from clinical
specimens. Int J Med Res Rev.
2015;3(8):866-870. doi:
10.17511/ijmrr.2015.i8.163.
- Kuti JL, Wang Q, Chen H, et al. Defining the
potency of amikacin against Escherichia
coli, Klebsiella pneumoniae, Pseudomonas
aeruginosa, and Acinetobacter
baumannii derived from Chinese hospitals
using CLSI and inhalation-based breakpoints. Infect
Drug Resist. 2018 May 25;11:783–90.
- Bora A, Sanjana R, Jha BK, et al. Incidence of
metallo-beta-lactamase producing clinical
isolates of Escherichia coli and Klebsiella
pneumoniae in central Nepal. BMC Res
Notes. 2014 Aug 21;7:557.Available at https://bmcresnotes.biomedcentral.com/articles/10.1186/1756-0500-7-557
- Brink AJ. Epidemiology of carbapenem-resistant
Gram-negative infections globally. Current
Opinion in Infectious Diseases. 2019
Dec;32(6):609-616.
|



















